Identification and characterization of immune correlates of tuberculosis disease and risk across species
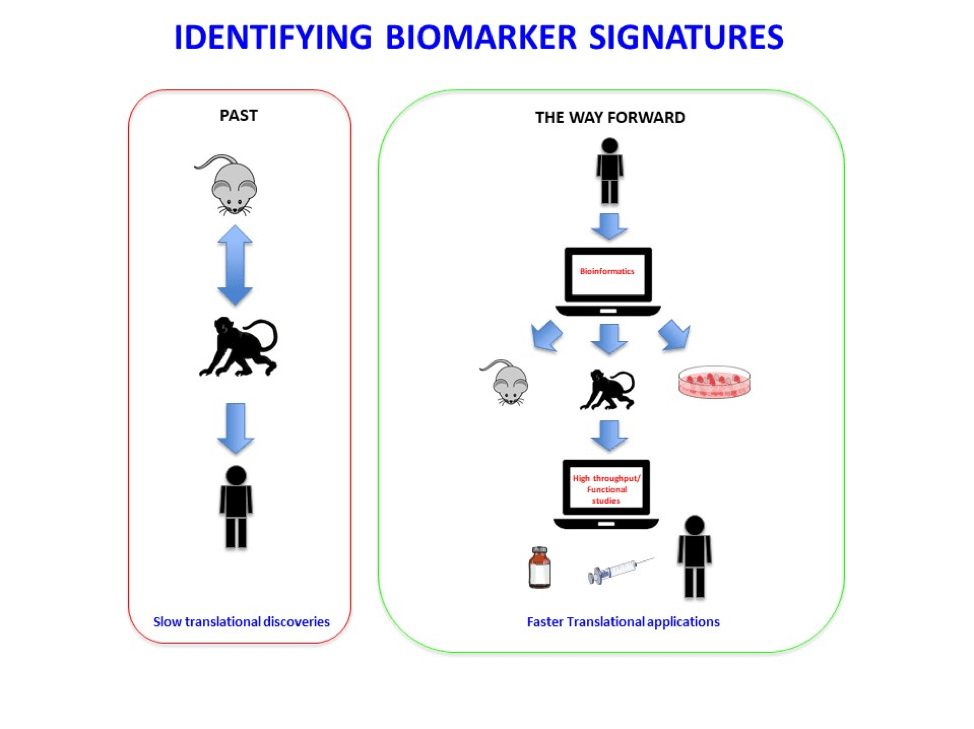
Animal models have been extensively used to characterize immune mechanisms of TB. However, immune correlates of protection or inflammation identified using animal models have not been validated in human studies. Immune correlates identified using animal models are not always effectively translated to human TB, thus resulting in a slow pace of translational discoveries from animal models to human TB for many platforms including vaccines, therapeutics, biomarkers and diagnostic discovery. Therefore, it is critical to improve our poor understanding of immune correlates of disease and protection that are shared across animal TB models and human TB. We have investigated the common immune correlates of risk of TB disease across animal models and humans and have mechanistically interrogated the functional role of these genes in Mtb infection using diversity outbred mouse model along with gene deficient mouse models. We have provided an in-depth, unbiased identification of the conserved and diversified gene/immune pathways in non-human primate and diversity outbred mouse TB models and human TB. The results from our studies show for the first time that in fact prominent differentially expressed genes/pathways induced during TB disease progression are conserved in genetically diverse mice, macaques and humans. Additionally, using gene deficient mouse models, we have addressed the functional role of individual genes comprising the gene signature of disease progression seen in humans with Mtb infection. Our data show that genes representing specific immune pathways can be protective, detrimental or redundant in controlling Mtb infection, and translate into identifying new immune pathways that mediate TB immunopathology in humans (Ahmed et al. 2020). Together, our cross species findings provide novel insights into modeling TB disease and the immunological basis of TB disease progression.