Research
The Prince Lab’s research program has evolved and expanded from an initial fascination with the regionalization of developing vertebrate animals. This interest led to a focus on the Hox genes, which play a major role in conferring regional identity. Using the zebrafish as our primary model, we took a deep dive into the organization, function and evolution of Hox genes, studies that provided important insights into whole genome duplications in the vertebrate lineage, as well as the patterning of neuronal and neural crest cell populations. We also made an extensive foray into regionalization of the endoderm, especially the endocrine pancreas. Currently, we are once again focusing investigations on the endlessly fascinating neural crest as well as studying the evolution and development of the anterior lateral line sensory system. Collective cell migrations have also been a common theme in our studies — of facial branchiomotor neurons, of neural crest, and now of the developing pronephros. Find out more about our projects below and by clicking on individual lab members.

The neural crest
We are interested in exploring the mechanisms that underlie development of the neural crest (NC), a remarkable vertebrate-specific cell type. NC cells are a transient, migratory cell population that gives rise to a large array of cell types, including neurons, chondrocytes, and pigment cells. NC cells are initially specified at the border between the neural and non-neural ectoderm. We are especially intrigued by neural crest regionalization and have found that the homeobox gene cdx4 plays a critical role in regulating posterior neural crest. Our ongoing studies of the zebrafish neural crest are focused on fate mapping, exploring the role of neural crest in anterior lateral line patterning, and investigating the control of cell fate decisions.

Neuronal migration
The Prince lab has a long-standing interest in the mechanisms that control migration of neurons. We have used the facial branchiomotor neurons (FBMNs) of the zebrafish VIIth cranial nerve as a model system to investigate this question. Zebrafish FBMNs are born in rhombomere (r)4 of the hindbrain before 16 hours post fertilization (hpf), and their cell bodies migrate in a chain-like manner towards the posterior. Our research has shown that FBMN migration depends on the function of Hoxb1a (a Hox patterning gene expressed in a dramatic “stripe” in r4), as well as on its downstream effector Pk1b. Although Pk1b is typically thought of as a planar cell polarity molecule, we have shown that Pk1b also functions as a nuclear translocator of RE1-silencing transcription factor (Rest). In turn, Rest is needed to maintain the immature state of the neurons during migration. We have also investigated the cellular basis of FBMN migration, using ablation and confocal imaging strategies to describe the “pioneer” neuron that leads migration through the hindbrain. Our most recent studies used photoconvertible transgenic tools and Light Sheet Microscopy to unravel the relationship between FBMNs and octavolateral efferent neurons. To learn more about this work, check out Vicky’s presentation from the April 2021 British Society for Developmental Biology Meeting, shared on the left.
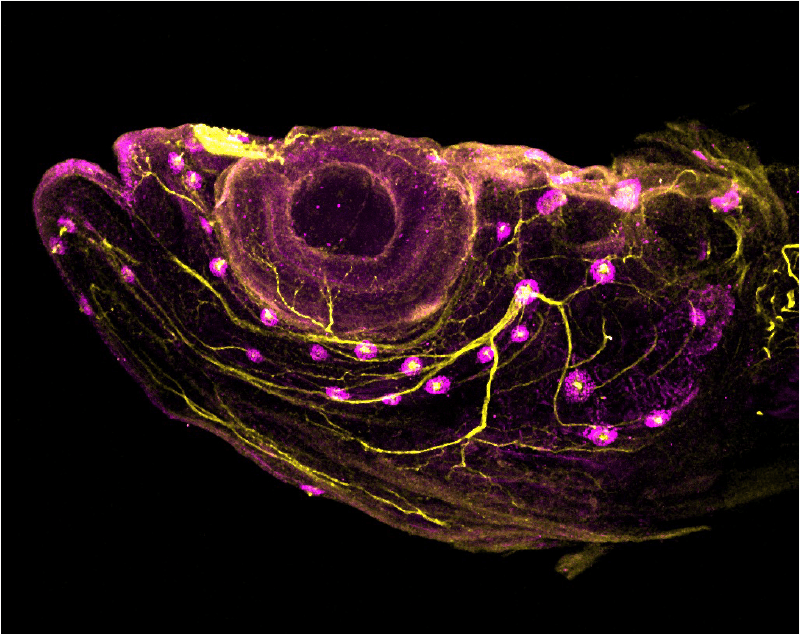
Evolution and development of the anterior lateral line
We are studying the anterior lateral line sensory system in not only zebrafish, but also in little skate and other species. The lateral line operates only in aquatic vertebrates and consists of mechanosensory hair cells housed in structures called neuromasts, embedded in canals or grooves —or lying superficially —over both head and trunk. The neuromasts are innervated from the lateral line ganglia, to provide aquatic organisms with a sense of ‘touch at a distance’, critical for schooling behavior and prey or predator detection. While we know a great deal about the development of the posterior lateral line, we know much less about the anterior lateral line, which branches around the eyes and across the jaw and cheeks. We are investigating anterior lateral line development using a range of approaches including light sheet imaging of transgenic zebrafish specimens, immunolabeling, and microCT scanning (in collaboration with the Coates lab). Ongoing projects focus on how anterior lateral line patterning is influenced by adjacent neural crest; innervation of the expanding lateral line system—including at postembryonic stages; and the placodal origins of skate lateral lines (in collaboration with the Gillis lab).