Splicing Catalysis
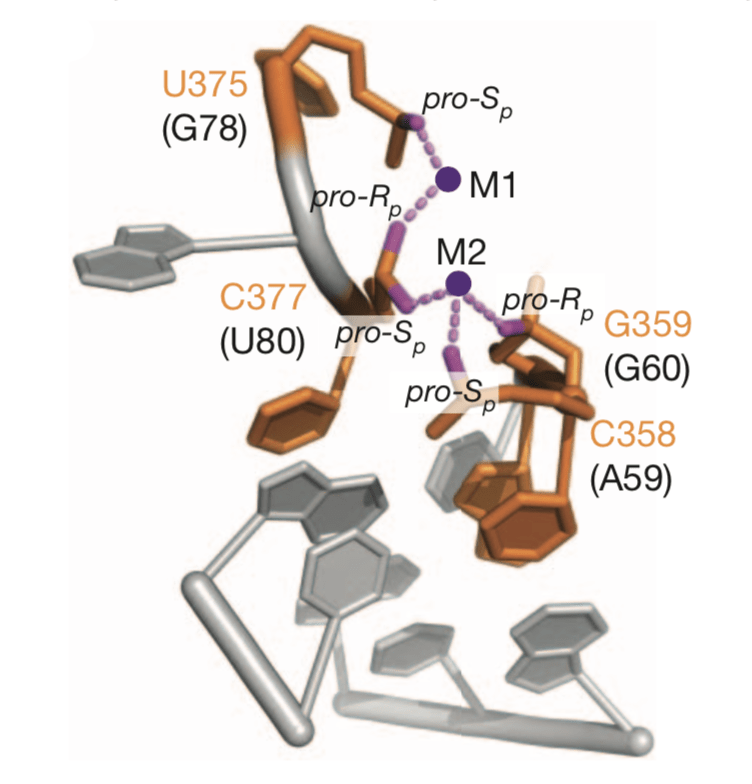
Pioneering work by our lab, in collaboration with Joe Piccirilli, shows U6 snRNA coordinates metals to cataylze both transesterification steps of pre-mRNA splicing, further supporting the hypothesis that the spliceosome and the group II self-splicing intron share an evolutionary origin (Fica & Tuttle et al., Nature, 2013).
Mechanisms of DEAH-box Helicase Translocation
Through the combination of x-ray crystallography, yeast genetics, and comparative structure analyses, we have uncovered the importance of several structural motifs in mediating the 3′ to 5′ translocase activity of the model DEAH-box helicase Prp43 (He et al., RNA, 2017).
Genome-Wide Interrogation of Splicing
Using the model organism S. cerevisiae, we developed a method of enriching for and sequencing excised lariat introns, allowing interrogation and discovery of novel branch points and 5′ splice sites (Qin et al., RNA, 2016).

Alternative Splice Site Selection
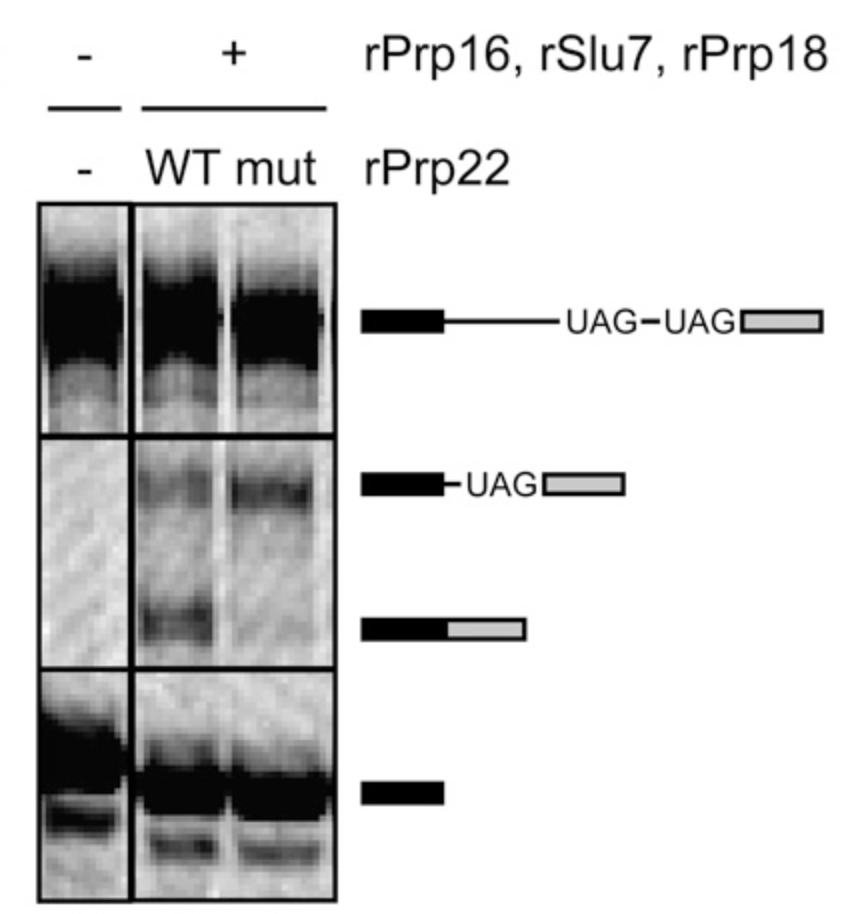
Alternative splicing is a major form of transcriptome diversity, with upwards of 90% of human genes being alternatively spliced. Our lab discovered both alternative branch site usage and alternative 3′ splice site usage can be mediated by the ATP-dependent activities of the DEAH-box helicases Prp16 and Prp22 (Semlow et al., Cell, 2016).
Fidelity of Splicing
As an essential part of gene expression, splicing has to proceed with high fidelity. Work from our lab shows DEAH-box helicases in the spliceosome work to proofread several steps in the splicing process (Semlow & Staley, TiBS, 2012), from Prp2 proofreading the catalytic core (Wlodaver & Staley, RNA, 2014), to Prp16 and Prp43 cooperating to discard spliceosomes stalled 5′ splice site cleavage (Koodathingal et al., Mol Cell, 2010), to Prp22 proofreading the exon ligation stage of splicing (Mayas et al., NSMB, 2006).
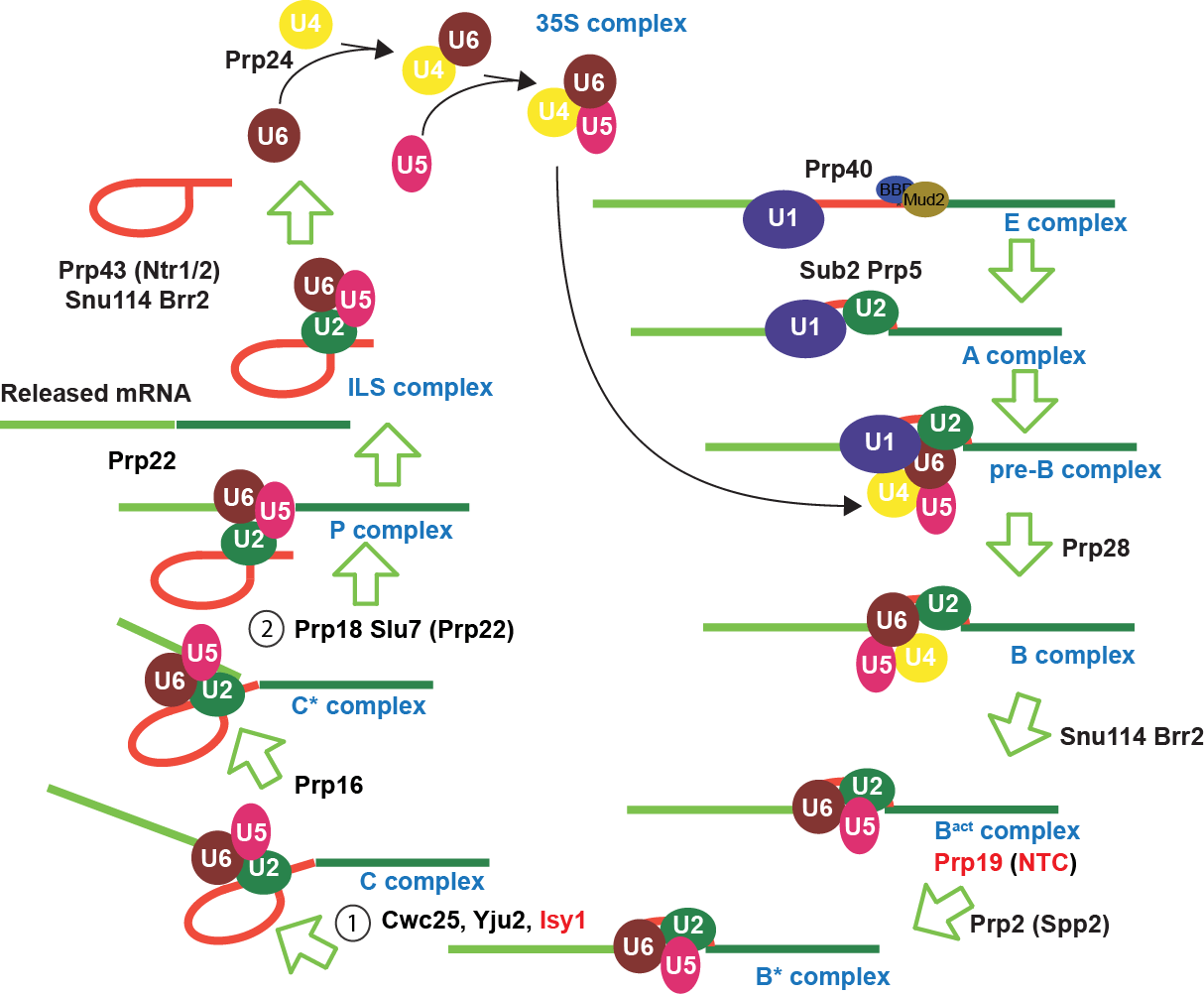
Spliceosome Dynamics
Unlike its cousin the ribosome, the spliceosome is inordinately dynamic, with several large-scale rearrangements that occur during assembly, activation, catalysis, and disassembly. Central to these rearrangements are Superfamily-2 helicases. Our lab shows these helicases initiate RNA-RNA rearrangements necessary between the two chemical steps of splicing (Hilliker et al., Genes Dev, 2007; Mefford & Staley, RNA, 2009), as well as act to discard splicing intermediates (Mayas et al., PNAS, 2010). We’ve also shown misregulation of helicase activity can lead to diseases such as retinitis pigmentosa (Zhao & Bellur et al., Am J Hum Genet, 2009).