Using flow cytometry to assess cell death, and more specifically, apoptosis, is an everyday occurrence in a flow core. Technically speaking, performing the assay couldn’t be easier when utilizing numerous kits available from any number of vendors. It’s typically set up in the format, add ‘x’ ul of reagent A to your cells, then ‘x’ ul of reagent B, and analyze. So, with such a simple assay, what could possibly go wrong?… Lots!
Let’s assume there are no faults with setting up the instrument or parameter voltages and the like. Let’s also assume the assay worked; that is, control cells looked relatively live, and treated cells looked nicely dead. The possible problem I’ve been coyly hinting at therefore, is in the analysis.
What’s typically the first thing someone does when they analyze their flow data? They make a forward scatter (FS) versus side scatter (SS) plot and gate on the ‘live’ cells. Obviously, if you’re trying to determine frequencies of live and dead cells, you wouldn’t use a ‘live’ gate, you’d extend the gate lower on the FS parameter to include the dead cells. However, when you do so, you could be including small debris and bits of cells, and it’s not exactly fair to count fragments of cells as events. So, herein lies the dilemma: You’d like to count whole cells, while drawing a light scatter gate that encompasses both live and dead/dying cells. Hence, My 3-step approach to gating Annexin V data appropriately.
Step 1: Display your Annexin V channel versus your Membrane Permeability Channel (for ease of typing, I’ll assume we’re using Annexin V FITC and PI) on the entire, ungated data set, and draw a region around the double negative (DN) fraction.
Step 2: Using the DN region from Step 1, gate a FS versus SS plot on these DN events, and draw a tight region around the subset of cells with low FS, now called Debris.
Step 3: Invert this Debris gate, creating a Not-Debris gate, and use this Not-Debris gate on the total population for your downstream Annexin V vs. PI display.
To see a screencast walkthrough of the gating process, watch the video below:
Let me break this down for you using an example. The entire point of the 3-step exercise is to properly define an area of the FS vs. SS plot that you’ll call Debris, which will not be counted when generating your downstream frequencies. Depending on how much death you induce, and how toxic your treatment is, you could have a significant amount of Debris in your sample. It’s important to note that you’re not simply throwing away these events, you’re just removing them from the downstream classification as Live (Annexin V neg, PI neg) Early Apoptotic (Annexin V pos, PI neg), Late Apoptotic (Annexin V pos, PI pos), or ‘Necrotic’ (Annexin V neg, PI pos). These debris events belong to none of these categories and therefore should not be included. The harm in including these debris events is that they usually end up in the Live fraction since they neither have a nucleus nor an intact membrane. Therefore, leaving them in inflates the Live fraction of cells, yielding potentially misleading results. The figure below shows the respective plots and regions.




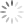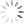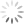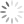
0 Comments