We are interested in the evolution of gene regulation in metazoans, a large, diverse and highly successful group of multicellular organisms.
Specifically, we aim to understand how genes can be wired into gene regulatory networks to perform their functions in distinct, tightly regulated contexts, a concept commonly referred to as pleiotropy. The evolution of pleiotropy required modifications to the genetic program that included the advent of specialized promoters and regulatory elements such as enhancers and repressors.
The key questions we address are the following:
1. What mechanisms do different animal lineages employ to tightly regulate where and when genes are expressed?
2. What modifications to the genome at the level of chromatin organization have occurred to facilitate changes in gene regulation?
3. How are individual gene expression programs integrated at the level of the cell to give rise to distinct phenotypes?
Our current understanding of gene regulation is limited to a small number of vertebrate and invertebrate genetic models. Advances in systems genomics allows us to examine regulatory mechanisms in phylogenetically relevant taxa.
Single Cell Genomics
To develop a quantitative framework that allows us to measure where and when genes are deployed within the context of a developing animal, we utilize single-cell RNA-seq approaches. These technologies allow us to determine how genes are used during development and homeostasis across all tissue and cell types. We are also able to utilize this data to design reporters for in vivo tracking of cell types.
Flora Plessier
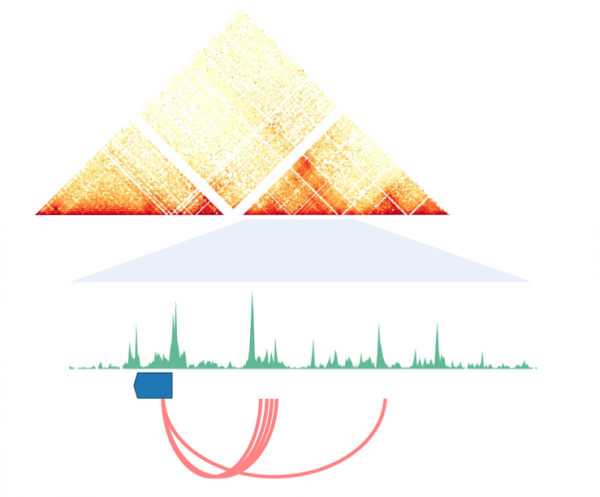
Gene Regulation is Facilitated by Chromatin Organization
Animal genomes are folded within the limited 3D space of the nucleus. The organization of this folded chromatin and its regulation by chromatin-interacting proteins enable the operation of the cell-type specific regulatory programs required to guide the developmental process. 3D chromatin landscapes, much like the linear genetic sequence, are subject to evolutionary change. These lineage-specific modifications have important implications for the ways in which animal genes are transcriptionally regulated and, we hypothesize, the evolution of morphological complexity.
Jelena Scepanovic