Our laboratory has a long-standing interest in B cell antigen receptor (BCR) signaling and how BCR dependent processes regulate specific cell fate decisions and contribute to disease pathogenesis.
The Molecular Mechanisms of B Cell Development
In the bone marrow, we have been working to understand how signals initiated through the pre-BCR, in conjunction with those delivered through the IL-7 receptor, coordinate cell cycle progression with immunoglobulin light chain gene recombination. These studies resulted in discovery of the epigenetic reader BRWD1 as critical for both regulating Igk accessibility and in coordinating broad transcriptional programs in early and late B lymphopoiesis. Subsequent studies have demonstrated the importance of BRWD1 in human immunity and in peripheral adaptive immune responses. We have also demonstrated that CXCR4 signaling directly Igk recombination. These latter findings fundamentally rewrite the canonical model of B lymphopoiesis and challenge current paradigms of chemokine receptor function. In our studies, we have developed novel in vivo and in vitro models to obtain definitive insights into the fundamental molecular mechanisms of lymphopoiesis.
The Role of B Cells in Autoimmunity
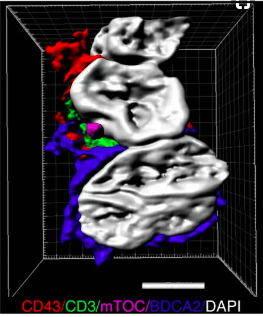
Our human translational studies have focused on in situ adaptive immune responses in humans and how they contribute to both autoimmunity and cancer. For these studies, we have used deep machine learning to develop methods that can be applied to human tissue samples to identify when any antigen presenting cell population is presenting peptides to T cells. Remarkably, this bioinformatics platform, which we term Cell Distance Mapping (CDM), approaches the sensitivity and specificity of two-photon excitation microscopy (TPEM). However, unlike TPEM, it can be applied to the study of human disease. We are now using CDM to quantify adaptive cell networks across several autoimmune diseases. In addition to quantitative imaging, we also use single cell technologies and high-throughput antibody expression to understand B cell selection at sites of inflammation and determine the interrelationships between transcriptional state and antigenic specificity. Overall, our goal is to obtain a functional understanding of how human adaptive immunity evolves at sites of disease.