Research in the Piccirilli Lab
The central theme of the Piccirilli Lab is RNA biochemistry and structural biology. The lab is especially interested in RNA catalysis and non-coding RNAs. Currently, our research spans ribozymes, riboswitches, mechanisms of splicing, siRNA delivery in cells. We have pioneered the technique of Chaperone-Assisted RNA Crystallography (CARC), which involved in-vitro selection of Fragments of Antibody binding RNA, which promises a pipeline for efficient RNA crystallography. We are also actively pursuing mechanistic studies on the Spliceosome- a piece of RNA-protein machinery which carries out pre-mRNA splicing in eukaryotes. In collaboration with the Harris Lab at Case Western Reserve University, we are trying to elucidate the detailed mechanism of catalysis by the Hepatitis Delta Virus (HDV) ribozyme using Kinetic Isopote effects. In collaboration with the Lengyel Lab, we are developing techniques for detecting malignant tumors in Ovarian Cancer, using receptor-mediated internalization of imaging agents.
Structural and biochemical studies on ribozymes - Varied Satellite (VS) and HDV
Varkud Satellite (VS) Ribozyme is the largest nucleolytic ribozyme found in nature, which in its trans-acting form is only the third example of a multi-turnover RNA enzyme (after ribosome and RNase P). It eluded crystallization for over two decades, till our lab successfully solved the crystal structure of this ribozyme, which provided high-resolution structural evidence for the considerable amount of biochemical data accumulated over this period of 20 odd years. Further structural studies on this system have brought out interesting features about VS catalysis and its implications in the context of the RNA World. Hepatitis Delta Virus (HDV) Ribozyme is another nucleolytic ribozyme found in nature. We for the first time established the role of nucleobase mediated catalysis in ribozymes using HDV as a model system. Further thermodynamic and Kinetic studies have been done in our lab and the current efforts include mechanistic studies using Kinetic Isotope effects in HDV catalysis (In collaboration with the Harris Lab at Case Western Reserve University).
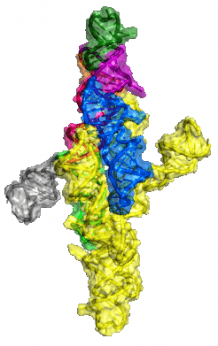
Crystal structure of the VS ribozyme
Structural and biochemical studies on the Spliceosome
Nuclear pre-mRNA splicing is catalyzed by the spliceosome, a massive RNA-protein complex. Our lab, in collaboration with the Staley lab, recently demonstrated that one of the RNA components of the spliceosome, U6 snRNA, catalyzes the two steps of splicing. We are now using the techniques developed in that project to probe other mechanistic aspects of splicing.
Publications related to this project:
Li NS, Tuttle N, Staley JP, Piccirilli JA. Synthesis and incorporation of the phosphoramidite derivative of 2′-O-photocaged 3′-s-thioguanosine into oligoribonucleotides: substrate for probing the mechanism of RNA catalysis. J Org Chem. 2014 Apr 18;79(8):3647-52
Fica SM, Mefford MA, Piccirilli JA, Staley JP. Evidence for a group II intron-like catalytic triplex in the spliceosome. Nat Struct Mol Biol. 2014 May;21(5):464-71
Fica SM, Tuttle N, Novak T, Li NS, Lu J, Koodathingal P, Dai Q, Staley JP, Piccirilli JA. RNA catalyses nuclear pre-mRNA splicing. Nature. 2013 Nov 14;503(7475):229-34
Koodathingal P, Novak T, Piccirilli JA, Staley JP. The DEAH box ATPases Prp16 and Prp43 cooperate to proofread 5′ splice site cleavage during pre-mRNA splicing. Mol Cell. 2010 Aug 13;39(3):385-95.
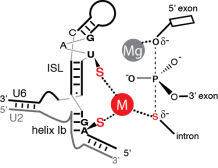
RNA catalysis in the spliceosome core
Protein Engineering, Synthetic Antibodies and Chaperone-Assisted RNA Crystallography
The Piccirilli lab is currently using phage-display technology for in vitro selection of RNA-binding Fabs. This method involves the bio-panning of specific nucleic acid structures against a Fab library where the phage particles consist of the Fab library fused to the gp3 coat protein. The library is generated by randomizing the light- and heavy-chain complementarity determining regions yielding more than 1010 different Fab variants. Selected Fabs can then be used in a variety of applications, such as chaperones for RNA crystallography, in vivo RNA pull-down assays, and diagnostic tools. Current targets for nucleic acid crystallography include ribozymes, riboswitches, aptamers, and microRNAs. In addition, the lab has focused on developing new synthetic libraries that substantially advance the selection of RNA antigens.
Publications related to this project:
Shao Y*, Huang H*, Qin D, Li NS, Koide A, Staley JP, Koide S, Kossiakoff AA, Piccirilli JA. Specific Recognition of a Single-stranded RNA Sequence by a Synthetic Antibody Fragment. (*Equal Contribution) J Mol Biol. 2016 Sep 2.
Huang H, Suslov NB, Li NS, Shelke SA, Evans ME, Koldobskaya Y, Rice PA, Piccirilli JA. A G-quadruplex-containing RNA activates fluorescence in a GFP-like fluorophore. Nat Chem Biol. 2014 Jun 22.
Koldobskaya, Y., Duguid, EM, Shechner, DM, Suslov, NB, Ye, J-D., Sidhu, SS, Bartel, DP, Koide, S., Kossiakoff, AA, Piccirilli, JA. A portable RNA sequence whose recognition by a synthetic antibody facilitates structural determination. Nat. Struct. Mol. Biol. 2011 18(1): 100-106.
Shechner DM, Grant RA, Bagby SC, Koldobskaya Y, Piccirilli JA, Bartel DP.Crystal structure of the catalytic core of an RNA-polymerase ribozyme. Science. 2009 Nov 27;326(5957):1271-5.
Ye, J-D, Tereshko, V, Frederiksen, JK, Koide, , Fellouse, FA, Sidhu, SS, Koide, S., Kossiakoff, AA, and Piccirilli, JA. Synthetic antibodies for specific recognition and crystallization of structured RNA. Proc Natl Acad Sci U S A 105:82-87 (2008).

Crystal structure of ‘Spinach’ bound to a synthetic Fab

