Our RESEARCH
Structural and mechanistic investigation of amyloid peptide-degrading proteases
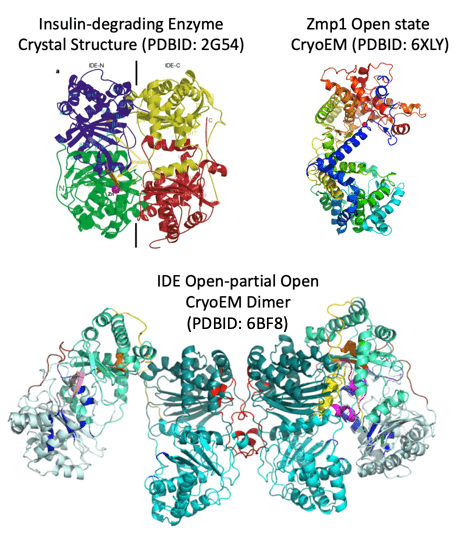
We’ve used an integrative structural approach to understand these crypt-containing peptide proteases from the M13 and M16 enzyme families.
2021 – Structural analysis of Mycobacterium tuberculosis M13 metalloprotease Zmp1 open states
2018 – Ensemble cryoEM elucidates the mechanism of insulin capture and degradtation by human insulin degrading enzyme
2014 – Anti-diabetic activity of insulin-degrading enzyme inhibitors mediated by multiple hormones
2006 – Structures of human insulin-degrading enzyme reveal a new substrate recognition mechanism

ExoY
Our results indicate that ExoY represents a new class of actin-binding proteins that modulate the actin cytoskeleton both directly, via F-actin bundling, and indirectly, via actin-activated nucleotidyl cyclase activity.
Key Publications:
2020 – Pseudomonas aeruginosa exoenzyme Y directly bundles actin filaments
Molecular Dynamics
All-atom and coarse-grained molecular simulations help us understand the dynamic nature of the cryptidase mechanism of action. We are using these technologies to guide our efforts to design cryptidase variants with engineered substrate specificity.


