We’re excited to announce that our NIH grant was officially funded, supporting our continued exploration of insulin-degrading enzyme through a variety of structural and dynamic investigational tools. Abstract and RePORTER link are below:
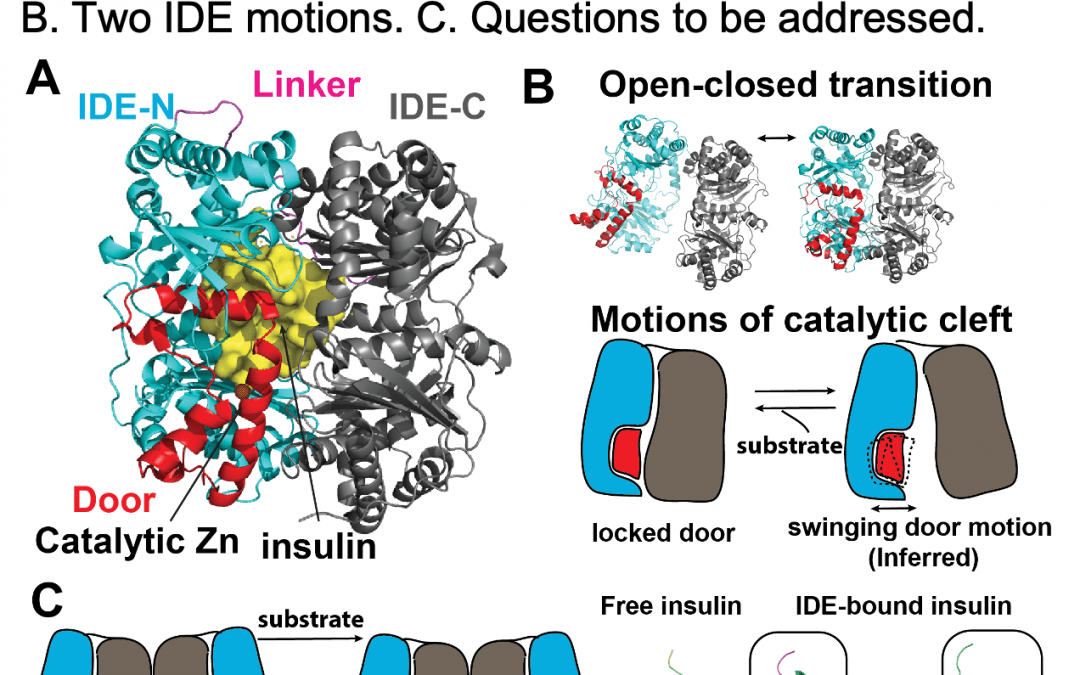
Project Abstract/Summary: Aggregates of amyloid peptides, such as amyloid fibrils are highly cytotoxic, as exemplified by the role of amyloid b (Aβ) in Alzheimer’s disease. To maintain a healthy proteome, a number of proteases target the monomeric form of amyloid peptides because this form fuels both seeding and elongation of amyloid fibrils. Insulin degrading enzyme (IDE) is a 110 kDa metalloprotease that degrades various amyloid peptides, including Aβ and three blood glucose-regulating hormones, namely insulin, amylin, and glucagon. Defects in IDE alter the progression of type 2 diabetes mellitus and Alzheimer’s disease in animal models and are linked to these diseases in humans. IDE inhibitors can control blood glucose level in mice and hold promise for treating diabetes. One of the key steps in the IDE catalytic cycle is the selective recognition and unfolding of amyloid peptides prior to degradation. Our premise is that the understudied conformational dynamics of IDE provide the mechanical basis for the unfolding of peptide substrates. Thus, we can leverage our understanding of these processes to selectively modulate the activity of IDE towards specific substrates. Our long-term goals are to elucidate the molecular details of how IDE selectively recognizes amyloid peptides and utilize this knowledge to develop novel IDE- based therapies to improve the human condition. Toward this goal, we have integrated ensemble structural determination and solution-based methods to show that IDE is a member of the chamber-containing protease, aka cryptidase, family that uses a sizable catalytic chamber to engulf monomeric amyloid peptides. We have also generated a working model that explains how IDE uses two key conformational switches to selectively degrade amyloid peptides. Our objectives for this application are to determine key unsolved conformational states and probe the conformational dynamics of IDE during the catalytic cycle by applying state-of-art integrative structural approaches. We will then combine MD simulation and screening to identify strategies to modulate the catalytic activity and selectivity of IDE. Our research rationale is that a deeper understanding of the regulation and functions of IDE will allow us to modulate its activity through engineering or novel small molecules and ultimately facilitate the design of IDE-based therapies to combat proteostatic imbalances. We will use time- resolved cryoEM and SAXS to understand the structural basis for substrate recognition during the key time window when IDE first encounters substrate in combination with advanced cryoEM image processing algorithms and MD simulation to address how IDE motions can unfold physiologically relevant substrates. We will apply the knowledge gained from the substrate recognition and unfolding studies to develop a screening strategy to identify methods to selectively modulate the degradation of Ab by IDE. This work will significantly enhance our understanding of the IDE catalytic cycle by defining key conformational states under physiologically relevant conditions and offer a platform to merge integrative structural analysis and MD simulation towards the discovery of innovative enzyme modulating strategies as the developmental foundation of novel IDE-based therapies.
https://reporter.nih.gov/search/1ZfH6qWGCUiOhF38aWxMSA/project-details/10367488#similar-Projects

Woo!