Jul 20, 2020 | Bioinformatics, Microbiome
by Elise Wachspress
Have you ever tried to follow a friend during a major marathon, like the one in Chicago?
This is only possible if you create a visible marker for either the runner, or—even better—the spectator, making it possible to identify each other in the huge crowds involved.
A red bandana is not going to cut it. You need something distinctive, highly visible and unusual, like maybe a giant letter “Q” balloon aloft above the throng!
Now imagine that you are trying to identify bacteria in the huge community of microbes nestled in your gut. Perhaps you want to find the particular ones involved in causing your illness or helping your body fight cancer. How do you identify these? Not only are there millions of “runners” in there, but they are reproducing hourly, which means the next generation might mutate and change the very “markers” you are using to follow them.
Nearly fifty years ago, Carl Woese, along with his graduate student Mitch Sogin, now a distinguished senior scientist at UChicago’s Marine Biological Laboratory in Woods Hole, Massachusetts, pioneered the use of a particular gene, called 16S ribosomal RNA, as a marker for microbes. Not only was the gene specific enough to identify types of bacteria; it was so “conserved,” or reliably unchanged over generations, that it became important as a “molecular clock” that could distinguish how closely bacteria might have been related over evolutionary history.
Woese used 16SrRNA to identify the archaea, a formerly unrecognized branch on the tree of life (and a great detective story!) Microbiologists now use the marker as a rapid, inexpensive shortcut to microbial identification. It’s just one of the arrows in the quiver of Sayantoni (Sam) Mukhopadhyay and her team as they study bacterial samples at the Duchossois Family Institute’s Microbiome Metagenomics Platform.
Mukhopadhyay’s team also helps researchers and clinicians sort out components of the microbiome with a newer, much more sensitive technique known as “shotgun metagenomics,” so called because of the somewhat scattershot way it targets pieces of the many genomes within a heterogeneous mixture. In this more complicated approach, the DNA from all the microbes in a sample is broken up into small segments and sequenced individually. After several rounds of fragmentation and sequencing, computer programs then identify the overlapping sections and reconstruct the sequences of the entire DNA strands from each bacterium.
This technique works especially well when some or all of the bacteria in the community have previously been grown in separate, pure colonies and fully sequenced by themselves. (Scientists and technicians have already laboriously sequenced the DNA of thousands of different bacteria and share these libraries.) These known bacteria become like the runners in a particular club who can always be identified by their specific combination of red bandana, white T-shirt, green shorts, and orange running shoes. Once these recognized sequences are accounted for, it’s easier to start homing in “new” bacteria DNA sequences not previously captured.
Not only can shotgun metagenomics provide an overview of complex communities of bacteria. It can also identify other DNA or RNA strands in a microbiome, like those in viruses—a benefit that seems clearer than ever in the time of coronavirus. Because shotgun metagenomics is so sensitive, the technique requires very careful preparation and handling of samples, so tiny DNA contaminants don’t skew the picture of what is actually going on in the microbial marathon.
And, as you might guess, the massive amounts of data involved require a lot of pretty sophisticated computational analysis. Eric Littman and Huaiying (Eddy) Lin are the seasoned bioinformaticians whose mathematical skills make sense of the endless combination of As, Cs, Gs, and Ts our internal athletes carry.
The new DFI Metagenomics Platform is a powerful tool for helping researchers and clinicians understand how microbial communities interact with each other and the human organism—information they are using here to understand how to resolve illness and, more importantly, promote health.

Jul 10, 2020 | COVID-19, Immunology
by Elise Wachspress with Peter Wang
Before COVID, before the Spanish flu, there was another highly infectious pulmonary disease that also changed entire countries and societies.
Tuberculosis, however, has been with us since antiquity. Like COVID, TB travels from person to person in droplets or aerosols. Caused by a mycobacterium, the disease was the most common cause of death in the nineteenth century. Populations who lived or worked in very close quarters—like migrants traveling in the holds of ships or poor people stacked in urban tenements—were the most vulnerable. But “consumption” was also somehow considered “romantic,” a wasting disease that also felled many writers and artists, the young John Keats famously among them. In fact, Edvard Munch’s “Sick Child,” above, documents his own sister dying of TB at age 15.
The advent of antibiotics made a big impact in curing the disease. But the mycobacteria that cause TB have evolved resistance, and poverty is still endemic around the world. The sad fact is that the disease still kills a million and a half people every year. That’s why the Bill and Melinda Gates Foundation is interested in TB research. One of the scientists whose work they support is UChicago’s Luis Barreiro, PhD.
Before antibiotics, French scientists Albert Calmette and Camille Guerin had already developed a vaccine against TB. In a way, it was similar to how the smallpox vaccine was developed from the much-less-virulent cowpox: Calmette and Guerin developed a vaccine for TB from a much less lethal bacterium also found in cows.
But BCG works unlike other vaccines. Most train the adaptive immune system, the specialized, “intelligent” commandoes that home in on very specific molecular targets. BCG also seems to affect the body’s innate immune system, a generalized assembly of cellular fighters once thought to be pretty untrainable. One clue is that BCG given in infancy or early childhood seems to reduce vulnerability to a whole range of diseases, not just TB.
And people now suspect that COVID-19 may be one of these.
Barreiro was working on TB and BCG long before the coronavirus reared its ugly head. He is developing the evidence that BCG ends up in the bone marrow, where it changes the epigenetics—the chemical “decorations” that hang off the basic structures of DNA and RNA—of the stem cells developing there. It creates something like a boot camp where newly emerging macrophages—the infantry grunts that rush to the front as part of the innate immune system—become better fighters against multiple diseases at once.
So the Gates Foundation is funding Barreiro’s research to understand whether this mechanism works in humans as it does in mouse models. In low-to-middle-income countries with high rates of TB and other infections, BCG is still given in infancy or early childhood and is known to reduce neonatal mortality for all causes. The Gates Foundation, focused on reducing TB in those countries, wants to understand how BCG works and if that knowledge can help scientists develop an even better vaccine.
One of Barreiro’s special talents in this quest is his abilities with “single-cell” technologies, because you can’t really characterize how different macrophages are working by looking at an “average” of their epigenetics and the “average” interactions they have with intruders. Barreiro uses automated processes that can look at individual cells—a lot of them, very quickly—to generate a fine-grained picture of how BCG vaccination fosters epigenetic variations in certain macrophages that allow these cells to fight many diseases.
Maybe even COVID.
As several recent journals have reported, there is growing evidence that countries with national BCG childhood vaccination programs have a much lower incidence of severe disease and death from COVID. So by developing an understanding of how BCG works, Barreiro and his team may generate important clues in protecting people from our latest scourge.
Science can’t, shouldn’t, won’t sit still and wait to address the next crisis. The work on TB (done centuries ago) and single-cell technologies (perfected over the past few years) may provide the answers to a crisis we didn’t even know we’d have to face.
Elise Wachspress is a senior communications strategist for the University of Chicago Medicine & Biological Sciences Development office. Peter Wang is a second-year undergraduate student in The College.

Jul 2, 2020 | COVID-19, Vaccination
By Elise Wachspress with Tinyan Dada
If you haven’t been infected yet, congratulations, you’ve made it this far.
We’ve streamlined our grocery store visits, maintained a six-foot distance at all times, even bought or made patterned, reusable masks for the family. We have a COVID routine, but we’re still waiting for the magic switch that will make everything normal again.
Thousands of scientists and technicians around the world are working to develop vaccines—substances that train our immune cells to attack particular pathogens—to keep us from getting COVID-19. Scientists continue to find multiple strains of the SARS-CoV2 virus circulating in various populations; it is becoming clear we may need multiple vaccines. And how long will these protect us? Measles inoculation works pretty much forever, but that is a rarity.
Sadly, even the most effective vaccination programs won’t work for everyone: the very young, the old, and especially those with impaired immune systems, for whom they may even cause disease. Will these people have to live forever in a COVID-19 bubble?
Maybe not. If we can establish herd immunity—where enough of the population is resistant to the disease—all of us might be able to resume a normal life.
But how many is “enough”? In an article in Immunity last month, UChicago geneticist Luis B Barreiro, PhD and grad student Haley E Randolph lay out the many parameters affecting herd immunity. In the case of this novel coronavirus, potentially influenced by countless unknowns—social structure, population density and age, even the genetic vulnerabilities of particular ethnic groups—assessing the potential for herd immunity becomes particularly uncertain and complicated.
The starting point, says Barreiro and Randolph, is to identify the average number of new cases each infected person might cause. In a completely susceptible population, where no one has experienced the virus before, this average is designated as R0. But once people start becoming resistant—either by getting the disease or through a vaccine—the effective reproduction rate, Re, starts to go down. The goal of vaccination programs is to get Re down below one, meaning more people have the disease than are transmitting it, and the disease curve begins to arc toward zero.
In the case of COVID-19, R0 has so far been estimated across different populations to be anywhere from around two to nearly six. (All of these R0 estimates were generated of course, without anyone understanding whether or how many people without symptoms can transmit the disease.)
So, Barreiro and Randolph suggest, suppose we take an average of the average—this totally new pathogen is forcing everyone to make a lot of guesses—and settle on an R0 of three. Mathematically, that would mean that incidence of the infection would begin to decline when about 67 percent of the population was resistant, and we would have a start—no promises!—toward herd immunity.
Complicating these models further are super-spreader events, like when one person singing in a church choir inadvertently infected at least 52 people with the coronavirus. We know from experience with MERS or even measles that one person or a tiny group can sometimes drive an inordinate number of infections. We don’t yet know how common these events are with COVID-19, although we have now learned that forcefully expelled droplets are a major mode of transmission, so taking precautions like wearing a mask, singing only at home, or even speaking more softly can ostensibly reduce R0.
Another important unknown is how long antibodies to the coronavirus might last. A year? Two? Because this coronavirus is so new, we have little idea how long the resistance we build up—either from having the disease or getting a vaccine—might last. Either way, protecting the vulnerable will depend on maintaining herd immunity over time.
Barreiro and Randolph go on to explain how to assess the infection fatality rate—the proportion of infected people who die of the disease—a metric critical in assessing the cost to society. Without isolation strategies, they project that worldwide deaths could exceed 30 million. Of course, as was the case in Italy, timing is everything: the faster the infection rate builds, the less likely the health care system can care for all the infected, and the more people will die.
In summary, Barreiro and Randolph point out that herd immunity is likely to work in only concert with a viable vaccination strategy, spread broadly throughout the population. Sweden’s coronavirus strategy involved keeping restaurants and businesses open, hoping that if less vulnerable members of the population interacted out in the community, they would generate some degree of herd immunity. So far, only around six percent of Swedes have developed COVID-19 antibodies, leaving the rest of the population at risk for serious illness and death and Swedes persona non grata visitors to other European countries.
So, in the short term, we continue to embrace our COVID routine. It is not much fun, but it may save our lives and the lives of others.
Tinyan Dada is a second-year undergraduate student at UChicago.

Jul 1, 2020 | COVID-19, Microbiome, News Roundup
Local EMT likely the world’s first double lung transplant recipient to survive COVID-19 via convalescent plasma. It was likely the only option to beat the disease for this young cystic fibrosis survivor, who received both the transplant and the plasma at UChicago Medicine. (Chicago Tribune)
UChicago will help with an experimental national vaccine trial for COVID-19. The study, spearheaded locally by the University of Illinois at Chicago, aims to test at least 1,000 people here. (Chicago Sun-Times)
The US is making progress on a universal flu vaccine. COVID-19 raises the stakes for fighting another infectious viral respiratory disease. (Axios)
Immune cells and noncancerous neurons can drive the growth of brain tumors in neurofibromatosis. Targeting those immune cells can help slow tumor growth. (Futurity)
Certain bacteria in the human gut may be driving a common liver disease. Researchers discovered the strain while studying a Chinese man who became intoxicated—without drinking—after eating high-carbohydrate meals, a condition they call “autobrewery syndrome.” (Science)

Jun 8, 2020 | Bioinformatics, Immunology, Microbiome
by Elise Wachspress
If you hang out with cutting edge scientists, you might hear or see the word (the suffix? a crossword puzzle answer?) “omics.” What are omics?
More than likely you‘ve heard of genomics, the study of the structure, function, evolution, and mapping of genomes, the collection of all the DNA in each organism.
And perhaps you’ve heard of transcriptomics, the study of all the ways an organism’s DNA is “transcribed,” or written into smaller molecules, the RNAs. While the DNA provides the basic, relatively unchangeable blueprints, environmental needs in the cell prompt the activation of specific genes. It’s like the highway engineer who, using her part of the blueprint, stages and directs the construction of one of the on-ramps. She’s working in concert with the larger plan, but somewhat separately from those directing other parts of the project.
Then there’s proteomics, the study of the structure and function of all the proteins that carry out the business in the cells. If the genome is a blueprint, and the engineering crew the RNA, the proteins are the molecular machines and building blocks—the backhoes, drills, and concrete—used to carry out the design.
The newest of the ‘omics fields is metabolomics: the study of all the chemical outputs of our cells and every microorganism that lives in and on us. These molecules, taken together, reflect the entire, functioning system, like how the cars and trucks using the highway are moving and thus creating new capital for society. Metabolomics is something like Here or Google maps, measuring important indicators of how the system is performing at both the street and system level. In a biological system, a genome can tell you what is possible, but the metabolome tells you what is actually happening.
But these readouts are more complex and critically useful: how a particular drug is working, how our immune system is responding, how microbes inside us are contributing to our health or modifying their environment to outcompete others, or even how our brains are prompting our bodies to act, and vice versa. New metabolomics technologies can help scientists non-invasively identify disease biomarkers, discover microbial products that can become new drugs, and identify the safest, most efficient ways to maintain health.
Among the many important resources the Duchossois Family Institute (DFI) is developing at UChicago is a facility that specializes in metabolomics. Led by Jean-Luc Chaubard, the DFI Host-Microbe Metabolomics Facility will feature state-of-the-art mass spectrometry, a powerful analytical technique that can be used to detail the profile of complex mixtures, whether solid, liquid, or gas. With this and other advanced instrumentation, DFI scientists will be able to understand the balance of molecules in blood, plasma, saliva, fecal, and even tissue samples, as well as in the waste products left behind when microbes are cultured (grown) outside the body.
Chaubard and his group will be looking at many things: from neurotransmitters and amino acids to bile acids and short-chain fatty acids, recently identified as critical to a healthy immune system. They will use large chemical libraries to create specialized “panels” that can profile multiple metabolites simultaneously. With these capabilities on campus, individual investigators will have ready access to new assays tailored specifically to their work.
The Metabolomics Facility will also help DFI investigators hone experimental design, decide when and how best to collect and store samples, and prepare those samples for testing. Importantly, they will also help in the data analysis that is critical in massive data-collection regimens like mass spec.
Chaubard, with a background both in academia (at Memorial Sloan Kettering and Caltech) and business (as founding director of the Molecular Discovery Lab at Modern Meadow, in New Jersey) is up for the challenge. An entrepreneur at heart, he is excited to be launching a resource that will set up UChicago as a leader in studying the convergence of immunology, the microbiome, and human health: “Here at UChicago, I get to work with some of the best scientists and doctors in the world, translating their work into practical applications that range from mechanistic understanding of human biology to early disease detection and discovery of novel drugs. It’s an honor to have this opportunity to improve the human condition.”
The Metabolomics Center is just one of the new platform resources made possible by a $100 million gift from The Duchossois Family Foundation and Craig and Janet Duchossois. We will bring you descriptions of several more in the weeks to come.
Elise Wachspress is a senior communications strategist for the University of Chicago Medicine & Biological Sciences Development office.

Jun 1, 2020 | Immunology
by Peter Wang and Elise Wachspress
How does COVID-19 pervert the very mechanism designed to destroy it?
Our bodies contain magnificent networks of cells, signals, and interactions that serve as sentries against harmful bacteria and viruses. But in the case of the coronavirus, it seems the most lethal blow sometimes comes from the immune system itself.
When a virus or bacterium—or sometimes even an unrecognized substance—enters the cells of the body, our immune systems send out small proteins to signal “get the war on these aliens started.” These small proteins, called cytokines, then attach to the outside of the cells and rev up inflammation, heating up the environment in an attempt to rid the body of the offenders.
In the case of this novel coronavirus, the response to the interlopers can quickly escalate, with more and more cytokines produced—a “cytokine storm.” It’s kind of like sending thousands of soldiers and tanks trampling across a landscape when the actual enemy is a guerilla cadre hiding safely underground: the potential result is a lot of damage done without ever defanging the real enemy.
For decades, Thomas Gajewski, MD, PhD, has led investigations of how to help the body fine-tune immune responses to cancer. In the coronavirus emergency that has commandeered all our lives, Gajewski is now working to understand the mechanisms of the immune response against SARS-CoV-2 in an effort to tamp down what may be the most dangerous element of this disease.
Within the first few weeks of the coronavirus pandemic in Chicago, Gajewski had put together a team of 40 investigators—from cancer clinicians to microbiome specialists to molecular engineers—to focus on detailing the mechanisms when the immune response to COVID-19 goes haywire. They organized a clinical study of 600 people—500 confirmed to have the disease and 100 healthy controls—and started measuring lots of parameters on each: the patients’ DNA sequences, blood cell composition, cytokines in the serum, antibodies and T cells against the virus, airway microbiota. They were looking for any relevant factor that shaped the immune response and potentially led to severe disease—the kind requiring intensive care unit support—versus mild disease that resolved spontaneously.
The goal: to understand which of these parameters was important in causing favorable or unfavorable outcome, and how clinicians could use these biological markers to decide the best treatment for each patient.
Already, Gajewski and his team had noticed that the lungs of some patients seemed to be full of macrophages, large white blood cells that protect places where our tissues meet the outside environment. And these macrophages seem to put out high levels of a specific cytokine known as interleukin-6 (IL-6), often implicated in severe cytokine storm.
Gajewski was well-positioned to launch this effort for multiple reasons. As a cancer immunologist, he has been studying how to increase or tamp down immune responses to disease for decades; he published his first papers on the interleukins nearly thirty years ago. To advance insights in cancer during decades of medical practice, he has relentlessly collected and carefully assessed as many patient samples as possible; these include samples less commonly studied, like stool—where he and his lab can identify the presence of the gut microbes that affect the immune response. And serendipitously, Jonathan Trujillo, MD, PhD, a fellow in Gajewski’s lab supported by Ruth and Elliot Sigal, had studied cytokines in coronaviruses earlier in his training, providing a direct expert on the team.
Gajewski and his team have already developed assays that reliably distinguish immune responses to SARS-CoV-2 and other coronaviruses. They are testing patients over a period of several days to follow disease progress and considering lots of questions. Does the degree of the immune response correlate with the severity of the disease? What other biomarkers correlate? Are there early warning signs that indicate which patients are most at risk for bad outcomes? Why does the drug Remdesivir seem to reduce the course of the disease? Does it also reduce the immune response?
With all the data the team is collecting, from microbial sequencing to patients’ personal genomes, they hope to develop much more knowledge about COVID-19 and why certain patients seem to get much sicker, in a very short period of time. The multiplex tests they are developing—assessing many types of cytokines and other markers at once—will also be extremely useful in addressing other diseases.
Cytokine storms are a complication not only of COVID-19 but of respiratory diseases caused by other coronaviruses, such as SARS and MERS. In fact, an out-of-control cytokine response was linked to the high fatality rate for the 2005 outbreak of the H5N1 “bird flu” virus. They are also associated with non-infectious diseases, such as multiple sclerosis and pancreatitis, and are a common side effect of the cancer immunotherapy approach utilizing CAR-T cells. So what we learn about these mechanisms can help advance medical science on several fronts.
Peter Wang is a second-year undergraduate student in The College.
Elise Wachspress is a senior communications strategist for the University of Chicago Medicine & Biological Sciences Development office.

May 29, 2020 | Microbiome, News Roundup
UChicago immunologist Cathy Nagler is developing immunotherapies for food allergies.
Work in her lab has launched a company focused on resetting the balance of beneficial bacteria in the gut to reduce the danger of foods like peanut butter. (Scientific American and Knowable Magazine)
Dodging the coronavirus is still the primary microbial concern of most Americans.
Chicago epidemiologist Emily Landon’s rules of thumb: outside beats inside, mask better than no mask, smaller groups rather than larger. (National Public Radio)
Maintaining a healthy microbiome during COVID-19 may be more important than ever.
Some clinicians in China have suggested that COVID-19 patients with gastrointestinal symptoms might be experiencing worse outcomes from the virus. (The Conversation and Gut-British Medical Journal)
Statins may work to improve the gut microbiome in patients with obesity.
Statins appear to increase gut diversity and reduce inflammation in these individuals. (Genetic Engineering & Biotechnology News)
Environmental contaminants strongly influence gut microbiome health.
Scientists at the University of Illinois at Urbana-Champaign have linked dozens of chemicals, including bisphenols and phthalates (plastic food packaging); PCBs and perfluorochemicals (nonstick cookware); and pesticides and herbicides, to changes in the gut microbiome and health issues. (Technology Networks)
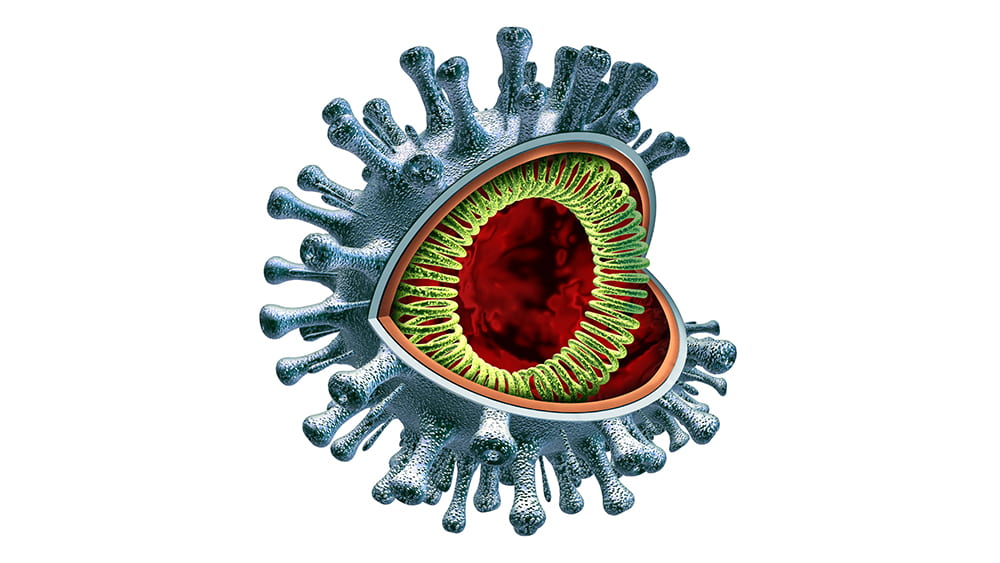
May 13, 2020 | Immunology, Vaccination
by Roma Shah
In this COVID-19 pandemic, the need for virus research has never been greater. People are dying, and right now there seems no effective way to stop it. In New York City, the situation has been overwhelming. Experts suggest this tragedy isn’t over; even if this first wave improves, the coronavirus may reappear, and by then we must understand its mechanisms, its manner of infection, and somehow create a solution.
Michaela Gack, PhD specializes in virology and immunology research, focusing on interactions between viruses and the immune systems of the hosts they infect. She breaks down this pandemic into three component areas: what it is, how people get it, and some ways researchers are looking to intervene.
COVID-19 is caused by a virus that belongs to a larger family of coronaviruses, including those that caused severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS), which emerged in 2003 and 2012 respectively. But the COVID-19 virus is a completely new version, and virologists have much to learn to stop this pandemic.
How do people get the disease? The virus, also called SARS-CoV-2, has proven to be highly transmissible, spread easily through the community, and we are just beginning to learn how. Complicating this problem is that a number of people who don’t show symptoms of infection apparently seem able to unknowingly spread the disease.
Although the virus—a very large piece of RNA encased in a protein capsule topped with a crown, or “corona” of spikes— is not “alive” by itself, it is extremely effective in using live cells for energy and spare parts to create many new versions of itself. By working to understand how it operates, Gack and virologists around the country hope to find ways—perhaps multiple ways—to intervene. They are considering several main opportunities.
The first is when the virus first hooks onto a human cell. It does so by attaching to an enzyme called ACE2 which is on the outer surfaces of the cells in the lungs and other organs. Imagine a locked door, where the virus must have the exact key to get into the cell. If scientists can find a way to jam that lock so the key can’t fit, then the cell will stay safely free of the virus.
One approach is “spike-binding”–creating an antibody that will clamp onto the virus’ “keys” and keep them from inserting into the lock. This is how a vaccine works, priming the immune system to make substances that lock onto the keys and keep them from spiking into the cell. But until scientists can develop that vaccine, we can gum up the keys by providing antibodies from patients who have successfully fought off the disease—what scientists call “passive immunity,” until we generate the vaccines to that will help patients to make their own.
Once the virus gains entry into the cell, it must next dissolve its protein coating and release its RNA payload. This is the point where some have suggested hydroxychloroquine might work. The drug, used to treat malaria and lupus, decreases the acidity inside cellular vesicles, working somewhat like a knot in a scarf, to keep the viral coating intact.
Once the RNA is released, it starts replicating uncontrollably, in an interesting way: it uses the building blocks inside the cell to generate long strands of RNA. An enzyme encoded by the virus called polymerase facilitates this process, adding those building blocks along until the chain is complete. Some of these RNA molecules are used to make constituent viral proteins, up to a million of them in each cell.
This is where the drug remdesivir has been hypothesized to work. Remdesivir very closely resembles one of the four main building blocks of RNA. So, when the polymerase is building the chain and looking for the RNA components, it could accidentally grab remdesivir instead, and the replication process falls apart. Think of using screws to put furniture together, where two look very similar. Pick the “wrong” one and the whole desk falls apart.
One more avenue for intervention is in editing the RNA molecules of the virus once they are in the cells. Cellular RNAs contain more than a hundred fifty important modifications at thousands of sites, some with critical regulatory roles analogous to those of protein and DNA modifications. UChicago researchers Chuan He and Tao Pan are world leaders in RNA modifications, and they are now working on ways to deactivate the RNA production that takes over the cellular machinery.
This COVID-19 pandemic, all its unknowns, and all these possible avenues for treatment remind us once again of the important role researchers play even in the most emergent situations, searching for results that can benefit our society as soon as possible.
Roma Shah is a second-year undergraduate studying neuroscience and public policy.

May 4, 2020 | Microbiome, News Roundup
Can plasma from patients who survived coronavirus help treat COVID-19 patients?
Clinical trials at UChicago Medicine are about to find out. (Chicago Tribune)
Did a UChicago researcher discover the first coronavirus?
In 1965, Dorothy Hamre took up the challenge to study students with colds and identified a new kind of virus. (Forbes)
A mutant enzyme that might solve a recycling crisis
Discovered in a compost heap, this enzyme can reduce bottles to chemical building blocks in a matter of hours. (The Guardian)
Do early solid foods predispose babies to be overweight?
Research from Johns Hopkins suggests changes in the microbiome in infancy can affect babies over a lifetime. (Futurity)
The breast is best, perhaps especially for preemies.
A study in newborn mice shows a component of breast milk may help protect premature babies from developing sepsis. (Futurity)

Apr 27, 2020 | Immunology, Research, Transplantation
by Peter Wang
When I started college at UChicago, I thought “This is going to be the best four years of my life! Don’t waste any of it!” I would try out for water polo, make lifelong friends, and jump into research. I wanted to become a doctor in the future and was eager to get my hands dirty.
The spring of my first year, I joined the lab of Maria-Luisa Alegre, MD, PhD. Her lab studies the responses of T-cells, the guardians of the body against foreign invaders, in solid organ transplantation: T-cell tolerance to donor grafts, the impact of infection and inflammation on anti-graft immunity, and interactions between the host immune system against the transplant and microbiota at different body sites.
Roughly 40,000 transplants were performed last year, and over 100,000 people are still on the national transplant waiting list. What the Alegre lab uncovers about the immune mechanisms involved in transplant rejection and tolerance has important implications for the health of these transplant recipients.
My first year as a scientist went by quickly, like when you’re driving somewhere and you lose your sense of time. I found my own groove, balancing chemistry classes in the morning with mouse experiments in the afternoon, not to mention lots and lots of pipetting. I felt I was doing something important, and life couldn’t be better.
Then my life—everybody’s lives—hit a massive speed bump. In March 2020, we were faced with the looming COVID-19 pandemic. My daily routine quickly shifted to life under lockdown: online classes, no labs, stress and anxiety and facing the anti-climactic reality that all I could do to help was to stay indoors. I had so much to look forward to that month; not only exciting experiments, but also a water polo tournament in Des Moines—vanished.
This pandemic has undoubtedly affected the lives of millions of students, small business owners, and brave healthcare workers on the front lines. But I get to give you my perspective, that of the cohort I am joining—scientists—the people best positioned to get us out of this mess.
Scientists in immunology are all motivated by the desire to understand health and disease and improve human health. The more we know about the immune system, the better outcomes we can provide for patients experiencing cancer, organ failure, infection, and allergy.
To advance knowledge, many immunologists utilize unique mouse models. In studying transplantation, Dr. Alegre suggests that “mice are relatively easy to alter genetically, offering the smallest animal model in which it is still possible to transplant an organ (if you are a skilled microsurgeon) and which allows mechanistic investigations into how a given immune gene can lead to graft rejection or graft tolerance. These genes can then become therapeutic targets and improve patient outcomes.”
In the Alegre lab, we use mouse models to study T-cell responses following skin and heart transplantation. Many of our mice are transgenic; like genetically altered fruits or vegetables, our mice have T-cells or tissues engineered to express or lack certain genes and proteins. We use these as donors or recipients of transplanted organs or as sources of transplant-reactive T cells to understand the immune response to the graft.
Because of the limitations in the number of complex surgeries that can be performed in a day, we often plan and perform multiple mouse experiments simultaneously. In her lab, Alegre says that “many of our experiments look at the maintenance phase of transplantation tolerance, 30-60 days after transplantation. Our microsurgeon usually generates a continuous stream of transplanted mice of the various gene backgrounds to have a lineup of experimental mice that have already reached the desired time point.”
Confronted with the necessity to evacuate the lab, we knew there would be few, if any, people left to care for the mice. Soon, universities and labs across the world would ask researchers to wrap up all ongoing experiments and think long and hard about the mice they needed, freeze the embryos of rare and special strains, prioritize the young pups of unique strains, and in many cases, cull the rest.
“The week in which we had to shut down the lab and dramatically reduce our mouse colony was distressing,” Alegre said. “In the span of a week, we erased years of work, prioritized irreplaceable strains and sacrificed all non-essential animals. Mentoring experimental design, methods and data interpretation is obviously much less effective when bench experiments cannot be performed, impacting undergraduate and graduate education.”
Our final in-person lab meeting was heartbreaking. We prioritized our mice: the young and pregnant were spared, with the hopes of ramping up breeding once the lab shutdown is over. Alegre estimates that this crisis has caused a “setback of about a year to be where we were before the lab shutdown. Publication of our findings will be delayed until we can ramp up transplantation and get to the point where we can repeat experiments and generate new data.”
As we adjust to this newfound reality, many at UChicago and the Duchossois Family Institute are confident that our life-saving work will continue, but at a different time and pace. When asked about the future, Alegre expressed that “we scientists are resilient by nature, however we’ll miss the thrill of new discovery and the quiet satisfaction of doing something useful that advances scientific knowledge.” We’ve shifted to lab meetings and journal clubs via Zoom, and I am writing about our work now in a different way—for the public rather than in scientific journals for other researchers. But the sacrifice and isolation in the age of social distancing is one of our important duties as citizens, and saving lives is what I value most.
Peter Wang is a second-year undergraduate student in The College.