Edit January 19, 2023: The 2021 Nature paper by Carlos P. Roca et al. introduced AutoSpill, which resolved the gating issue raised in this blogpost. In essence, AutoSpill uses iterations of linear regressions to get the best compensation or unmixing matrices, and removes the need for gating the positive and negative fractions of your controls. It saves you time, and generally provides better results. Learn more about AutoSpill here.
Edit October 21, 2021: It should be noted that the tips for gate placement described in this post can be applied to spectral unmixing as well, just replace the term “compensation” with “unmixing”. For more tips on spectral unmixing specifically, see this video.
When calculating compensation, automated tools are the gold standard. However, people often struggle to get good results from the automated compensation tools and will turn to manual compensation to fix any errors. Why is it difficult to get accurate results from automated tools? The only explanation that I’ve heard from the flow cytometry experts is that suboptimal results are caused by poor quality compensation controls. In this post I’d like to demonstrate that poor controls are not always the cause. The part that is often overlooked is that automated compensation tools are not fully automated – they require users to subjectively set gates on positive and negative cells within each control. Yes, some of the tools do have automated gate placement, but that feature often fails to generate appropriate gates. Gate placement is a critical component in determining the outcome of an automated compensation tool.
Setting Up the Automated Compensation Tool
Before I discuss gate placement I want to quickly go over how autocompensation tools work. Compensation determined to be correct when the median fluorescence intensity (MFI) of the positive population is in line with the MFI of the negative population (Figure 1). The flaw of automated compensation tools is that the user usually determines where to set the MFI for each population by placing gates on the positive and negative populations. Thus, the accuracy of the compensation is dependent on a subjective gate placement.
To demonstrate the effect of gate placement I’m using FlowJo’s automated compensation tool, though this should be applicable to any comparable software. I tested four different gate positions (Figure 2):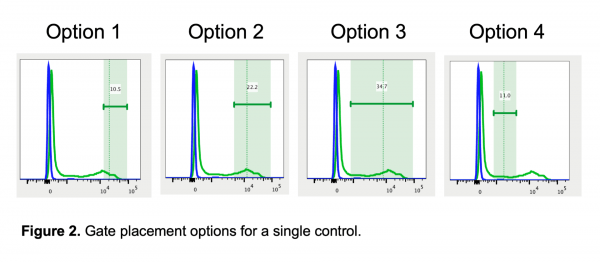
- The most positive (brightest) of the positive cells, which cuts through a high-expressing population and would never be used for analysis
- The whole bright/high-expressing population of the positive cells, assuming this control has a bright population and a dim population
- The entire positive population of cells, including dim and bright cells
- The whole dim/low-expressing population of cells, avoiding the bright/high-expressing cells
Results of Automated Compensation
To check the accuracy of the compensation determined by the automated compensation tool, I applied the compensation matrix to the same file and checked 5 plots. Again, the compensation is correct when the MFI of the positive and negative populations match. Here are my results:

The compensation using the first gate – on the most positive of the positive cells – gave the most accurate results. The second gate was nearly perfect, though two of the 5 plots are very slightly undercompensated. The compensation from the third and fourth gates produced suboptimal results with significant compensation errors. These data highlight that setting gates for autocompensation is very different from setting gates for analysis. The first gate would never be used to analyze data, yet it provides the best compensation results. The other gate options could be used for data analysis, but clearly do not work as well for calculating compensation.
Why does the first gate work so well to calculate compensation? If you remember the rules for compensation controls, one of the rules is that the control must be as bright or brighter than the sample. Compensation can be calculated for cells that are the same intensity or dimmer than the control, but calculating compensation for cells brighter than the control does not work well. Since compensation is calculated by MFI, that rule should be translated to state that the MFI of the control must be as bright or brighter than the sample. The first gate works the best for calculating compensation because the MFI (shown as the dashed vertical line in FlowJo in Figure 2) is the highest. In all the other gates, there are cells that are brighter than the MFI used to calculate the compensation, so those gates have essentially created a control that breaks the rules of a “good quality compensation control”.
In practice, my recommendation is to use an automated compensation tool, but know its limitations and always check the accuracy of the compensation prior to analyzing data. For best results, use high quality compensation controls and set the gates on the brightest cells. When setting the gates on the controls, remember that you are not analyzing the data, it is perfectly acceptable to cut a through a population with the goal of calculating a high MFI.
Looking for more flow cytometry resources?
Compensation
Learn how to choose between compensating on the cytometer or in an analysis software, tips for troubleshooting compensation errors, etc.
Spectral Unmixing
Learn tips and tricks for performing spectral unmixing in SpectroFlo, how the spectral unmixing algorithm works, etc.



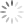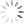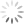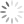
Well written article and clear explanation. Thanks for sharing Laura
Excellent article. Thank you.
I had been wondering about optimal gate placement for marking the positives for single color compensation controls. This article answered all my questions, and explained why. Thank you!
This is great! I love the images. How about placement of the negative gate when you have both populations in one tube? I’ve seen some folks draw gates differently depending on whether they’re using log or biex (and how appropriately the biex is scaled). I haven’t systematically tested it, but it looks like you have a good dataset for it!
Hi Rachael,
Good point! I used the universal negative for this dataset, and I haven’t played around with the negative gates much either. In general, I tend to put the gate on the most negative of the negative. I’ll try to play around with the negative gates and see if it makes any impact.
Laura
I think this also shows the importance of titrating antibodies on beads to get the brightest signal. I saw so many people using the exact same concentration on beads as what they have on cells.
hi i just want to clarify that it IS ok to cut through a pop for the compensation but you just wouldn’t do it on your sample or FMOs right?
Hi Ashley,
The best gate placement for setting up a compensation wizard is one that captures the brightest intensity, even if that cuts through a population. The best gate placement for analyzing a fully stained sample (assuming you’re interested in separating positive and negative cells) is to draw it based on an FMO control. You could gate through the middle of a “population” if the FMO justifies it (like when there is a continuous signal between negative and positive cells instead of two distinct populations). It is very likely that the gates needed for the compensation wizard are completely different than gates needed for sample analysis.
Once you understand the mathematics underlying compensation, you realize that setting the gate with the highest possible MFI of the positives is your best bet. Note – on FlowJo, you have to take at least 2% of the events in the histogram.
Mathematically, the data is correctly compensated when the MFI of the positives and the MFI of the negatives in the secondary detector(s) are equivalent, not just look like a straight line.
The blog and videos are well-explained. Learned a lot. Thank you, Laura.
Awesome article.
Thanks Laura, great job in explaining this..
Very helpful tip. Thanks Laura
Hello, i just came across this gem and I’m so grateful for this. Completely helped me understand compensation. I have one more question: what’s the best way to determine the appropriate voltage for FSC-A and SSC-A on a heterogenous sample? eg. heart sample. I’m struggling to find the right voltage that would lead to a good singlets gating, thank you
Hi Gregory,
Good question. First off, let’s clarify that your FSC and SSC voltages will not impact the compensation or unmixing matrix. All that you’re trying to do when setting up the voltage on your intrinsic parameters is to showcase your population of interest. Basically, you want to set it up pretty much in the middle of your figure. It can be challenging with complex tissues – like your heart tissue – but here are some tips to help out:
– Use SSC on log scale: the log scale squashes your data on a smaller portion of the axis, making it easier to identify the different clusters that may be present, and reducing the chances of loosing your more complex cells off-scale;
– Set a gate on the top bin of your FSC axis. That will allow to get a percentage value of the events falling off-scale. They should be mostly clumps, but if that percentage is high (say 10% and up), you may be losing cells of interest and could benefit from a lower FSC voltage value.
– One easy trick that will allow you to identify your cells of interest is to start your acquisition looking at a marker expressed on your cells of interest. You can gate on the positive events on that parameter, then using the backgating feature (using a color dot plot, for example), you can identify where that population of interest is falling on your FSCxSSC graph – they will be the colored dots that you can then display at the center of your graph.
– Regarding the singlet part of the question, I still think your FSC-A x FSC-H graph is the proper tool. Use the SSC-A x SSC-H to clean up some more if needed.
– Lastly, if you have access to specific instruments, you could make use of their special features. The siPM detectors on the Novocyte platforms makes it very unlikely that you will lose samples due to poor gain settings, so it’s a sure way to record your data. And if you have access to an Attune CytPix (or an ImageStream MarkII), you will be able to collect images of the events on your FSCx SSC graph and see what they actually are. This should give you better clarity on what you’re looking at.
Hope this helps! Let me know if you need clarifications!
Thank you so much for clarifying many of my doubts with this article.
I have been working with a big Panel of 11 colors. I tried to use compensation beads, but I failed. Now, after reading this article and all the replies I will try to fix the gates of compensation.
Nevertheless, I also used cells (single stained) for compensation too, in a different experiment for the same 11-color panel, and this time, the matrix of compensation looked better, but still, I can see that I had spillover in some of the channels, mostly among the yellow channels, maybe because the same problem with the gates for compensation.
I want your opinion about using beads or cells for compensation. What are the advantages or disadvantages?
And, is it possible to have a compensation matrix with high spillover numbers, like more than 300 or 600, and still see the cell population? I mean, the MFI aligned between the negative and positive populations.
Or should I always see the numbers of the spillover matrix lower than 100, for example?
Any recommendations or opinions are very welcome. Thank you so much.
Marlene
Hi Marlene! There has been some developments on this question of cells or beads in the last couple of years. I have another blogpost on that topic here – but in short, there is quite a number of fluorophores that will not provide correct correction when controls are prepared with beads. The hypothesis right now seems to be that the beads and the fluorophores are reacting to one another and as a result, the fluorphore signature is modified and no longer matches the one it would have on a cell. As a result, the unmixing/compensation is incorrect and there is no real solution to fix it. So at this time, the answer to the question cells or beads is a definite cells!
Regarding the matrix values: there is no ceiling to what they should be. If it takes a 300 to properly compensate, then that’s what you need. However, I would not expect this on a 11 color panel – I don’t know your panel or what instrument you’re working on, but on our 4 lasers platforms, I’d expect a choice of fluorophores that is manageable. In other words, you’ll have high matrix values if you have high overlap between your fluorophores, and should otherwise not be expecting them. So if you get these values, I’d look for whether or not you’re compensating on the actual sample as opposed to high autofluorescence or something like that. Secondly, I’d revisit the voltages (or gains) on your detectors to make sure they are all optimized.
I hope this helps, happy to discuss in more details.
Hello David,
Thank you so much for your answer and advice. This blog is gold for me!
About our flow cytometer. We have an Attune NTx with 4 lasers: Violet (4), Blue (3), Yellow (4), and Red (3). We have been using the Ultracomp Beads and working until now, maybe because we used panels of 4 or 5 colors. This is the first time I have tried to use a panel with 11 colors. This time the UltraComp beads are causing issues with the compensation.
I read your previous post about using beads or cells. And apparently, the UltraComp beads are not a good option. I am wondering if the flow cytometers used in the cited article are Cytek Aurora and the BD FACSAria Fusion are comparable with the Attune NTx?
Thank you so much for your help. Any other suggestions are welcome.
Marlene