When the global pandemic hit and Chicago’s shelter in place orders prevented me from going into the lab, I started doing some experiments in my kitchen instead. I decided to take on baking sourdough bread (like many other quarantined individuals). And while I learned to perfect my bread in my free time, I spent a lot of my working hours on training materials for new flow cytometrists – that’s when I started to realize some similarities between these two tasks. It’s been a while since I learned flow cytometry, but baking bread reminded me of the process of learning a new skill: follow a protocol, assess the results and identify problems, determine how to fix the problems, and repeat the protocol with adjustments to address the problems. As I created my flow cytometry training materials I realized that I didn’t cover all of these steps. Flow cytometry training materials usually focus on the protocol of how to perform a flow cytometry experiment, but they don’t always cover how to troubleshoot. The step that is often skipped is how to assess your results to identify problems. With baking this is easy for everyone, even a novice – you taste your food and immediately know it doesn’t taste good. But for novice flow cytometrists this step of identifying problems can be very difficult. Looking back, I realized that no one really ever told me what bad data looks like. It was just a skill that I eventually picked up. So in this post I’d like to go over some examples of bad data to assist novice flow cytometrists.
There are a lot of ways to get bad flow cytometry data – in this post I’ll focus on the reasons that I feel are the easiest to identify and fix. From this post, you should be able to identify if there are major issues with the instrument settings (which can only be fixed by re-recording the samples) or if the samples need to be cleaned up in analysis software with gates or a new compensation matrix.
FSC and SSC Settings

FSC and SSC should be set so that all of your cells of interest are within the plot. If FSC is set too low, it will be difficult to use the FSC to remove debris or noise. If FSC is too high, it can also be difficult to cleanly gate on single cells. The same rules apply for SSC. The example above shows a cell line with a distinct cell population. In this case, it should be relatively easy to set the FSC and SSC. However, tissue digests containing multiple cell types can be a bit more difficult if you are unfamiliar with the sample.

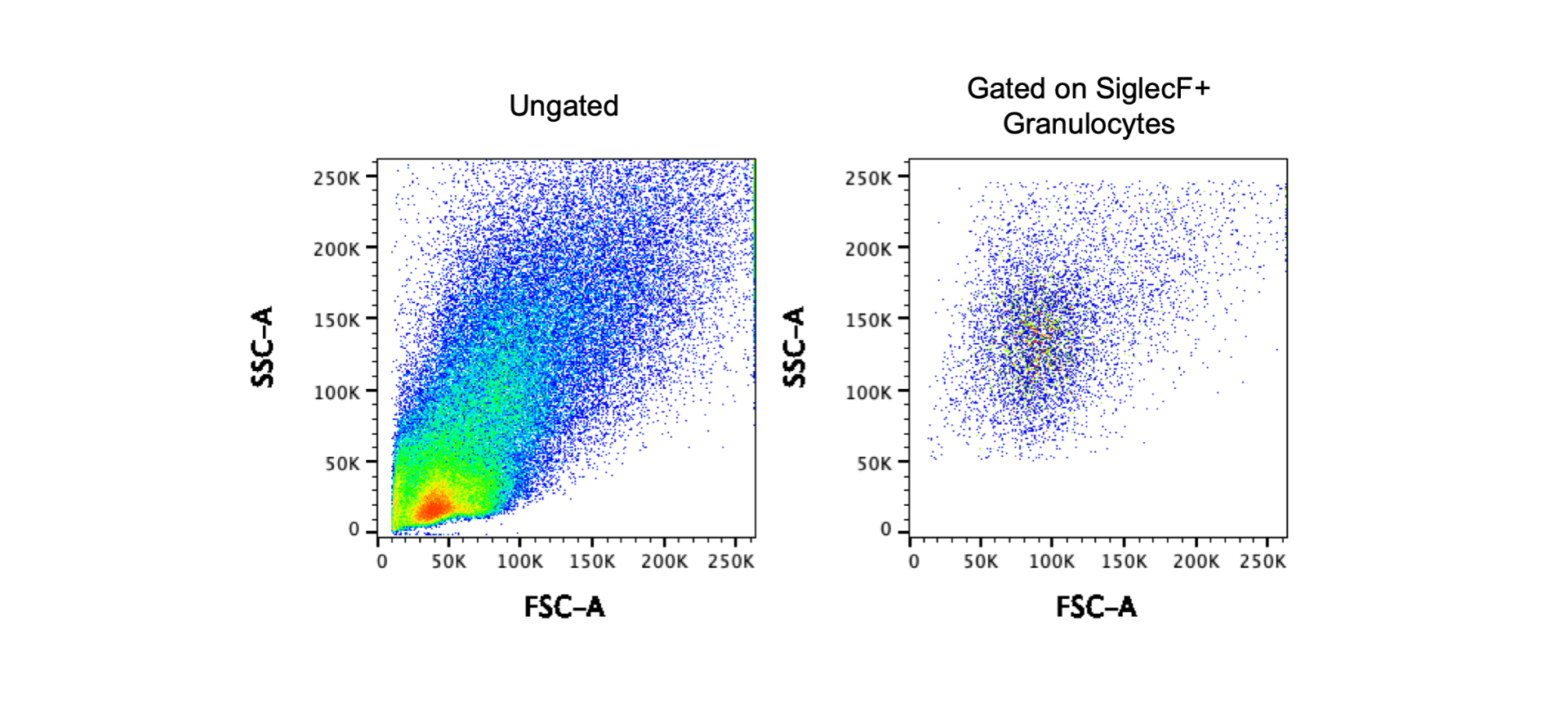
The above images show data from a tissue digest. The first plot shows the FSC and SSC for ungated cells and at first glance, these FSC and SSC voltages look great. However, I’ve mentioned that it can be more challenging to set FSC and SSC for digested tissue. To determine if the settings are correct, you can use a technique called backgating. In this case, I gated on CD4+ T cells or F4/80+CD11b+ granulocytes and then examined the FSC and SSC of the gated cells. The second plots shows that my T cell population is in the middle of the FSC SSC plot, but the third plot shows that the granulocyte population is on the top axis. In this scenario it is best to consider the goal of the experiment – if T cells or lymphocytes are the only cells of interest, these voltages work well. However, if the goal of the experiment is to look at granulocytes, the SSC should be decreased. The images below show a different experiment with different voltages using the same tissue. Using the backgating technique to first gate on SiglecF+ granulocytes and then look at the FSC and SSC, you can determine that these voltages for FSC and SSC are appropriate for this cell population.
Fluorophore Voltage Settings
For best results, avoid saturating the fluorophore detectors. Similar to the FSC and SSC, all fluorophore signals should be within the plot. The two examples below show data with incorrect voltage settings. Note that in the right plot, 71% of the events are saturating the detector.

Compensation Errors
Before proceeding with data analysis you should always make sure compensation is correct. My last blog post went into detail about compensation – read it here if you missed it! However a good flow cytometrist will always be on the lookout for compensation errors. You can easily identify compensation errors by looking at the negative portion of the axis – populations that are not symmetrical and below zero are concerning and can indicate compensation errors. The teardrop shape can sometimes be ok or it may be compensation or autofluorescence (third panel). However symmetrical spreading error is perfectly acceptable (read more on the trumpet effect here).

Antibody Aggregates
Another unusual pattern you may find in your data is caused by antibody aggregates. This pattern is a bit harder to identify because the flurorophores used to find the pattern are unique to the panel. However if you are finding a pattern of super bright events similar to the one below you may have antibody aggregates. For a completed data set you can remove them with a gate. To prevent them from happening you can centrifuge your antibodies for 10,000 RPM for 3 minutes prior to using.

Clogs and Other Issues with Cytometer Fluidics
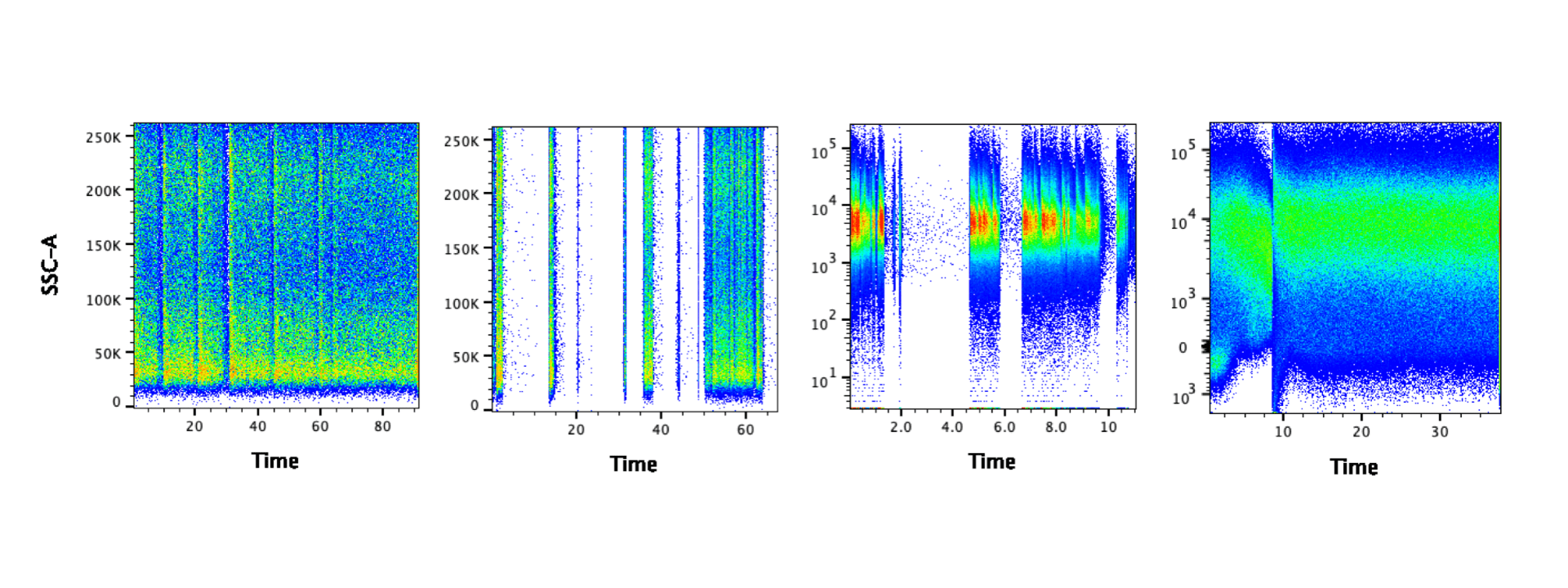
Before analyzing your data, it’s a good idea to look at the time parameter to examine the cytometer fluidics. If there were no issues with the fluidics, you should see an even signal for the duration of your sample. The images below show problematic data – the gaps suggest there may have been clogs. It may be possible to salvage the data by gating on the portion of data where the signal was steady. In some cases you can draw a gate manually – for example all of the data after 10 in the last panel below. Alternatively algorithms like FlowClean and FlowAI could also be used to clean the data. Please note: algorithms do not like slashes, so if any of the parameter names contain slashes, extra steps should be taken to remove them. If you are using FlowJo, you can use the export function to create new fcs files where all slashes are replaced with underscores. However if clogging is a recurring problem, you may consider filtering your samples prior to running them.
There is one situation where you will expect to find interruptions in the time parameter: data acquired by syringe injection. In this case, you will find the data is interrupted at regular intervals as the syringe empties and refills. Examples of cytometers with syringes include the Attune, BD fortessa HTS, and Helios. Any cytometers that require a specific tube so that it can be sealed and pressurized does not utilize a syringe. It is up to you if you want to remove the interruption of refilling the syringe, but it is recommended.
Summary
To set up the cytometer voltages/gains properly, all data (or as much data as possible) should be inside of the plot. Once data has been recorded, there is no way to alter voltages/gains. If it is determined that voltages/gains are incorrect, samples must be re-recorded on new settings.
Before analyzing data, you should check for correct compensation and inconsistencies in the time parameter. If compensation is incorrect, it is simple to generate a new compensation matrix to apply to the samples. If there are problems with the time parameter or if there are antibody aggregates, these can usually be solved during analysis with additional gating or a cleaning algorithm.
If you have questions about your data quality, I offer consultation services to help identify and troubleshoot problems. Also, this “bad data” post is the first in a series, stay tuned for the next one!
Update: Part 2 can be found here.
Looking for more flow cytometry resources?
Avoid Bad Data Series
Read the full series of blog posts.
Flow Basics 2.0
A comprehensive online course that covers the protocol and optimization of staining cells, panel design, choosing controls, and instrument setup.
Compensation
Learn how to choose between compensating on the cytometer or in an analysis software, tips for troubleshooting compensation errors, etc.




Great blog, I will be pointing my user base to this – very informative!!
Hi, great blog!
I have a question: I am working with dissociated brains and suffer a lot from autofluorescence, but cannot find the reason why, or what could eliminate it. Do you have any suggestions? And if it cannot be removed, any ideas on how to deal with it?
Thanks!
Good question! I think the autofluorescence of your cells are an intrinsic feature, I don’t think there’s a fun way to get rid of it. Cells from lungs have the same high autofluorescence which makes them somewhat harder to work with. There’s a couple of things you could do. If you have access to a spectral flow cytometer like the Aurora or the ID7000 from SONY, you should be able to subtract the autofluorescence during your analysis. If that is not a possibility, adjust your panel to avoid dim markers in the regions where autofluorescence is highest. Typically, the autofluorescence is higher in the low end of the spectrum. I’d put your bright, primary markers there. In the far-red region where autofluorescence should not be as big an issue, set your harder to resolve markers. Hope this helps!
This is quite helpful. Thank you so much!
I work with nerve tissues that are enzymatically dissociated before proceeding to FACS. I am interested in PNS-resident immune cells. As I am not trained in handling FACS machines yet, I usually submit my samples to our facility specialists and retrieve the raw data files for analysis (my analyses are almost always performed on the FACSAria III machine). When I try to analyse my samples, just as pointed in your tutorial, I notice my cells of interest, stained with BV605 (CD45+ population) are usually off-scale. When I try to plot the data on CD45 vs SSC-A, I get a ‘warning message’ that about 40% of the events are on the chart edges. When I gate populations and then backgate, to identify them in the FSC-SSC plot, the backgating shows that populations I have gated also include events from the bottom left corner of the dot plot. Do you think there is a problem with (a) FSC/SSC voltage (for the off-scaling problem) or perhaps PMT voltage settings for resolution/detection (b) threshold (otherwise why would events in the 0-50k axis of the FSC will be captured in supposedly CD45+ cell population).
Thanks in advance!
Hi Dami,
It’s always a challenge when someone else is running samples for you on a cytometer. If you don’t know how to direct the staff to set up the cytometer for your specific samples and if they make assumptions about your experiment instead of asking you then the end result isn’t always great. Having staff run easy samples (like cell lines or blood) is pretty straightforward, but it’s often quite difficult to get the correct voltage settings for digested tissue and it can take some trial and error to get the settings right.
Because of the situation you are in, I really think your best way forward would be to show your data to your facility specialists and discuss if/how they can make adjustments in the future. It’s difficult for me to give you a definitive answer without seeing the data, but from what you describe it sounds like you might have some issues with voltage settings. Your facility specialists should be able to help you determine if the voltage settings were correct if you provide them with your analysis and details about your tissue/experiment. If you’re seeing two CD45+ populations on FSC (large and small), then the smaller population might just be debris that needs to be gated out. Again, your facility specialists should be able to look at your data and and determine if the threshold setting is correct or not.
Hi,
we are running Purity and CD34+ using flow Cytometer
What could be why getting two different results, more than 30% different + failed from two preparation of the same sample, same run, same day, same reagent, and same instrument?
we checked the instrument before using CD-Chex 34, and everything was ok
we repeat the work using a fresh sample in different day we get the same result
Thanks
Hi,
This is not a question we’ll be able to answer without having a look at the data and the gating strategy.
Very well-written and informative post! I will be sure to let people I know who are confused with identifying bad cytometry data know about this one!
Great blog,
I have a question regarding negative fluoresence. I did a two color staining on a cell line, receptor staining with cy5 and a viability dye (e450). Even though I had live cells positive for cy5, there was still a significant population with negative fluorescence. Do you have any thought on what would cause this or how to fix it? Is there a way to shift the axis so that the whole population is positive?
Hi Elvin,
It hard to provide a good explanation without having a look at the data. Where is the negative fraction? On one parameter, both? Before you meddle with the data display, I would start by assuming that the measurements are correct and try to figure out what they mean. Do you expect all of your cells to be alive and positive for Cy5? And if not, what could be represented in the negative fraction? Debris, dead cells and such?
Do you have a negative control (unstained cells for example) that you can use to contrast. Do you have a positive control that will allow you to confirm the validity of your staining?
If you have a way to share the data (either a pdf of the fcs files), let us know.
Hi, I am recently working on mouse aorta tissue digestion and flow cytometry. Most of my collections have quite low FSC. Does that mean most of them are debris? Can debris be stained by antibodies? Will the viability dye stain the debris too?
Thank you!
Haiyan
Hi Haiyan,
Without having seen the data, please take whatever I’m about to say with many grains of salt. But yes, it does sound like your sample contains a lot of debris and dead cells – I’m basing this on the low FSC of the data. Those may contain the targets for your antibodies and could stain positive as well. Depending on your dead cell discriminator – wether a fixeable amine-based dye, or a DNA dye, you may get a positive signal as well. An easy way to make sure is to just get the sample under a microscope and have a look. Hope this helps!
Thank you, David! I used Zombie Aqua as viability.staining dye. I counted my cells under microscope which was stained with trypan blue. But the cell number via Flow doesn’t match what I counted with microscope. I mean I gated most of the cells but it is much lesser. For example, I counted about 5×10^4 small but round cells. I run all of them through Fortessa. I only have 5000 single cells. Does this sound right?
Hi Haiyan! No, I don’t think the discrepancy should be that high. I usually give a 10% variation to values coming from a microscope count, but we’re pretty far from it. If you have access to an Attune, a Novocyte instrument, or any other syringe platform that would allow you to measure the volume of acquired sample, use it to get a true count of your cells directly. Alternatively, counting beads are available at a (relatively) low cost.
Just to cover my bases, and because I’m curious about the 5000 figure, can you confirm that this is not just how many cells were recorded?.
It counts 22688 particles, I gated 11744 “cells”, singlet 8804, live cells 5561. Does this sound right?
Aloha Laura,
Thanks for posting this helpful information! I use flow cytometry to study algae in Hawaii – so we use autofluorescence for all our work. I used to be a cancer biologist at Feinberg having someone else run flow for me, but now I’m doing it myself on these unusual samples – pretty different work!
Hi John! Laura has moved to Cytek and is rocking it over there. We’ve been discussing imaging flow cytometry lately, and wether or not spectral offers just as good information. Please let me know of your publications, we’re getting very interested in that field. Cheers!
Hi there! I work in a lab doing flow cytometry, primarily leukemia/lymphoma evaluations. For our bone marrows we run 3 tubes with 10 markers each in them. One tube has T cell markers, one has B cell markers, and one has myeloid cell markers. We have a new staff who is getting significantly different lymphocyte populations for the same patient in the different tubes, and trying to figure out what could cause such an issue. Any ideas would be appreciated!
Hi Megan,
We discussed the issue, and presuming that your group has a standardized acquisition protocol on the instrument and an analysis template, we voted that your new person is making a mistake with the sample preparation. I don’t really have enough information to get a clearer picture, but other source of issues should be affecting your whole group.
Cheers!
Hello, in your first point you said the FSC was too low or too high however the values of the X-axis do not change. Could you please clarify? Thanks!
Hi Tiff! Laura was discussing the location of the clusters of events on the FSC axis. On the left panel, the clusters are all at the bottom of the FSC axis, which makes it harder to resolve the different population of cells that might be present in the sample. Increasing the FSC detector voltage would fix the issue. We have the opposite problem in the middle panel: a good chunk of the cell are stuck on the very top bin of the FSC axis. This tells us that the signal is saturating the detector and is likely caused by the FSC voltage setting set too high.
This is a good question. The FSCxSSC voltage settings is something that is not discussed in details in general. I’ll prepare a tutorial on the topic in the next few weeks, let me know if it helps!
Hi David! A tutorial on the FSCxSSC voltage settings would be great. When I look up my equipment operator manual (NovoCyte) it doesn’t seem as if there’s a procedure for adjusting the detector voltage. It would also be good to know what tips someone off to a detector voltage issue when looking at a population of cells (because you could simply adjust the ranges of X and Y to put your population in the middle of the plots). Thank you!
Hi
I was in the middle of a run and and I was prompt to refill the focusing fluid tank. Thereafter, my data went bad, the FITC and NF’s were close to zero, compared to the 2500 and higher. What could I have done wrong?
Hi Nnana,
This indicates that you either has so much noise being picked up that there was no chance of seeing your cells, or that the sample core in the stream was irremediably disrupted (due to bubbles in the flow cell or some other factor), or maybe more likely that air was still all over the system and the sample could not be pressured in the fluidic line. So the initial problem was to run an experiment with a close-to-empty sheath tank, I hope the lesson is learned here. Fill up every time. Following the problem, you need to purge the air out of the system and this is done in different ways for different instruments. We have some troubleshooting tips on the website that explains how to do that on the BD systems. Other platforms just won’t allow you to run the sheath tank dry. Once you have cleared the air out of the fluidic lines, it is worth your time running QC again just to make sure the instrument is back to normal.
Hello I have a question regardless of this article,
I was in trouble using the cytek
I conduct every experiment with the same concentration antibody. But It is not easy the size of plot is too big to gate. This problem is called antibody concentration. However, the same result continue even when the antibody is more added.
I think The problem is not Ab conc, but don’t know what the exact cause is
Hi Cho, I’m not sure I understand the issue you are experiencing. Yo can provide more details and I’ll see what we can do. This comment section doesn’t graph and images posting, so as an alternative I suggest you head for the Reddit or Discord Flow Cytometry and discuss your problems with the community over there. The links are here.